Abstract
Single-cell RNA sequencing (scRNA-seq) is a powerful tool for understanding cellular
heterogeneity and identifying cell types in virus-related research. However, direct identification of
SARS-CoV-2-infected cells at the single-cell level remains challenging, hindering the understanding of viral
pathogenesis and the development of effective treatments.
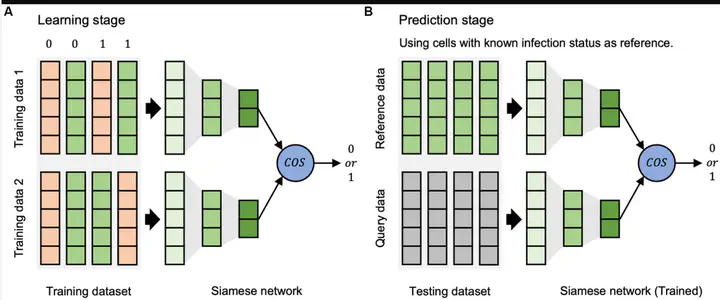
** Methods** In this study, we propose a deep
learning framework, the single-cell virus detection network (scVDN), to predict the infection status of single
cells. The scVDN is trained on scRNA-seq data from multiple nasal swab samples obtained from several
contributors with varying cell types. To objectively evaluate scVDN’s performance, we establish a model
evaluation framework suitable for real experimental data.Results and Discussion Our results demonstrate that
scVDN outperforms four state-of-the-art machine learning models in identifying SARS-CoV-2-infected cells, even
with extremely imbalanced labels in real data. Specifically, scVDN achieves a perfect AUC score of 1 in four
cell types. Our findings have important implications for advancing virus research and improving public health
by enabling the identification of virus-infected cells at the single-cell level, which is critical for
diagnosing and treating viral infections. The scVDN framework can be applied to other single-cell
virus-related studies, and we make all source code and datasets publicly available on GitHub at https://github.com/studentiz/scvdn.